GitHub - yihui-he/u-net: U-Net: Convolutional Networks for Biomedical Image Segmentation
Deep Learning Tutorial for Kaggle Ultrasound Nerve Segmentation competition, using Keras
This tutorial shows how to use Keras library to build deep neural network for ultrasound image nerve segmentation. More info on this Kaggle competition can be found on https://www.kaggle.com/c/ultrasound-nerve-segmentation.
This deep neural network achieves ~0.57 score on the leaderboard based on test images, and can be a good staring point for further, more serious approaches.
The architecture was inspired by U-Net: Convolutional Networks for Biomedical Image Segmentation.
Provided data is processed by data.py script. This script just loads the images and saves them into NumPy binary format files .npy for faster loading later.
The images are not pre-processed in any way, except resizing to 64 x 80. Since the images are pretty noisy, I expect that some thoughtful pre-processing could yield better performance of the model.
Output images (masks) are scaled to [0, 1] interval.
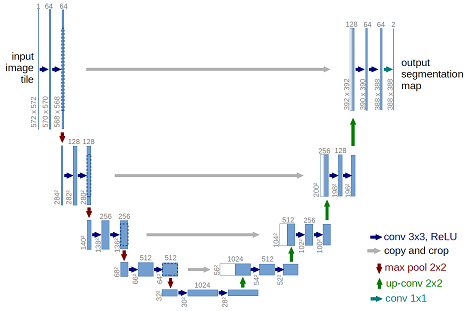
The provided model is basically a convolutional auto-encoder, but with a twist - it has skip connections from encoder layers to decoder layers that are on the same “level”. See picture below (note that image size and numbers of convolutional filters in this tutorial differs from the original U-Net architecture).

This deep neural network is implemented with Keras functional API, which makes it extremely easy to experiment with different interesting architectures.
Output from the network is a 64 x 80 which represents mask that should be learned. Sigmoid activation function makes sure that mask pixels are in [0, 1] range.
The model is trained for 20 epochs, where each epoch took ~30 seconds on Titan X. Memory footprint of the model is ~800MB.
After 20 epochs, calculated Dice coefficient is ~0.68, which yielded ~0.57 score on leaderboard, so obviously this model overfits (cross-validation pull requests anyone? ;)).